Input interpretation
(deoxy)ribose phosphate degradation (human metabolic pathway)
Pathway topology
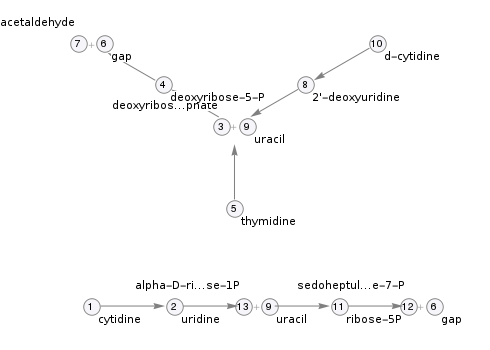
Pathway topology
Pathway components
![| cytidine | 1-B-ribofuranosylcytosine | 4-amino-1-[(2R, 3R, 4S, 5R)-3, 4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2-one | 4-amino-1-[(2R, 3R, 4S, 5R)-3, 4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]pyrimidin-2-one | 4-amino-1-[(2R, 3R, 4S, 5R)-3, 4-dihydroxy-5-methylol-tetrahydrofuran-2-yl]pyrimidin-2-one | 4-amino-1-beta-D-ribofuranosyl-2(1H)-pyrimidinone | cytidine free base | cytosine-1-β-D-ribofuranoside | cytosine riboside | cytosineβ-D-riboside | 243.2 g/mol | | C_9H_13N_3O_5 | uridine | 1-beta-D-ribofuranosyl-2, 4(1H, 3H)-pyrimidinedione | 1-beta-D-ribofuranosyluracil | 2, 4(1H, 3H)-pyrimidinedione-1 | 244.2 g/mol | | C_9H_12N_2O_6 | deoxyribose-1-phosphate | 2-deoxy-D-ribose-1-phosphate | 2-deoxy-alpha-D-ribose 1-phosphate | 214.1 g/mol | | C_5H_11O_7P_1 | deoxyribose-5-phosphate | 2-deoxy-D-ribose-5-phosphate | deoxyribose-5-P | 2-deoxyribose-5-P | 2-deoxyribose-5-phosphate | 2-deoxy-alpha-D-ribose 5-phosphate | 214.1 g/mol | | C_5H_11O_7P_1 | thymidine | 5-methyldeoxyuridine | deoxythymidin | deoxythymidine | 242.2 g/mol | | C_10H_14N_2O_5 | glyceraldehyde-3-phosphate | 170.1 g/mol | | C_3H_7O_6P | acetaldehyde | acetic aldehyde | aldehyde | ethanal | ethyl aldehyde | 44.1 g/mol | | CH_3CHO | 2'-deoxyuridine | 2'-desoxyuridine | deoxyribose uracil | deoxyuridine | uridine, 2'-deoxy | uridine, 2'-deoxy- | 228.2 g/mol | | C_9H_12N_2O_5 | uracil | 1H-pyrimidine-2, 4-dione | 2, 4(1H, 3H)-pyrimidinedione | 2, 4-dihydroxypyrimidine | 2, 4-dioxopyrimidine | 2, 4-pyrimidinediol | 2, 4-pyrimidinedione | hybar x | pirod | pyrod | 112.1 g/mol | | C_4H_4N_2O_2 | 2'-deoxycytidine | cytosine deoxyribonucleoside | cytosine deoxyriboside | deoxycytidine | deoxyribose cytidine | 227.2 g/mol | | C_9H_13N_3O_4 | ribose-5-phosphate | 232.1 g/mol | | C_5H_13O_8P | D-sedoheptulose-7-phosphate | D-Sedoheptulose-7-P | sedoheptulose-7-phosphate | heptulose-7-phosphate | sedoheptulose-7-P | 290.2 g/mol | | C_7H_15O_10P_1 | ribose-1-phosphate | 230.1 g/mol | | C_5H_11O_8P](../image_source/8b477092c275b9fb56bbc210dad8352b.png)
| cytidine | 1-B-ribofuranosylcytosine | 4-amino-1-[(2R, 3R, 4S, 5R)-3, 4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2-one | 4-amino-1-[(2R, 3R, 4S, 5R)-3, 4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]pyrimidin-2-one | 4-amino-1-[(2R, 3R, 4S, 5R)-3, 4-dihydroxy-5-methylol-tetrahydrofuran-2-yl]pyrimidin-2-one | 4-amino-1-beta-D-ribofuranosyl-2(1H)-pyrimidinone | cytidine free base | cytosine-1-β-D-ribofuranoside | cytosine riboside | cytosineβ-D-riboside | 243.2 g/mol | | C_9H_13N_3O_5 | uridine | 1-beta-D-ribofuranosyl-2, 4(1H, 3H)-pyrimidinedione | 1-beta-D-ribofuranosyluracil | 2, 4(1H, 3H)-pyrimidinedione-1 | 244.2 g/mol | | C_9H_12N_2O_6 | deoxyribose-1-phosphate | 2-deoxy-D-ribose-1-phosphate | 2-deoxy-alpha-D-ribose 1-phosphate | 214.1 g/mol | | C_5H_11O_7P_1 | deoxyribose-5-phosphate | 2-deoxy-D-ribose-5-phosphate | deoxyribose-5-P | 2-deoxyribose-5-P | 2-deoxyribose-5-phosphate | 2-deoxy-alpha-D-ribose 5-phosphate | 214.1 g/mol | | C_5H_11O_7P_1 | thymidine | 5-methyldeoxyuridine | deoxythymidin | deoxythymidine | 242.2 g/mol | | C_10H_14N_2O_5 | glyceraldehyde-3-phosphate | 170.1 g/mol | | C_3H_7O_6P | acetaldehyde | acetic aldehyde | aldehyde | ethanal | ethyl aldehyde | 44.1 g/mol | | CH_3CHO | 2'-deoxyuridine | 2'-desoxyuridine | deoxyribose uracil | deoxyuridine | uridine, 2'-deoxy | uridine, 2'-deoxy- | 228.2 g/mol | | C_9H_12N_2O_5 | uracil | 1H-pyrimidine-2, 4-dione | 2, 4(1H, 3H)-pyrimidinedione | 2, 4-dihydroxypyrimidine | 2, 4-dioxopyrimidine | 2, 4-pyrimidinediol | 2, 4-pyrimidinedione | hybar x | pirod | pyrod | 112.1 g/mol | | C_4H_4N_2O_2 | 2'-deoxycytidine | cytosine deoxyribonucleoside | cytosine deoxyriboside | deoxycytidine | deoxyribose cytidine | 227.2 g/mol | | C_9H_13N_3O_4 | ribose-5-phosphate | 232.1 g/mol | | C_5H_13O_8P | D-sedoheptulose-7-phosphate | D-Sedoheptulose-7-P | sedoheptulose-7-phosphate | heptulose-7-phosphate | sedoheptulose-7-P | 290.2 g/mol | | C_7H_15O_10P_1 | ribose-1-phosphate | 230.1 g/mol | | C_5H_11O_8P
Pathway steps

| reaction ⟶ | + H_2O ⟶ + ammonia | activation-induced cytidine deaminase | cytidine deaminase ⟶ | + H_2O ⟶ + ammonia | activation-induced cytidine deaminase | cytidine deaminase ⟶ + | + D-xylulose-5-phosphate ⟶ + | transketolase-like 1 | transketolase ⟶ | ⟶ | (none) ⟶ | ⟶ | (none) ⟶ + | ⟶ + | putative deoxyribose-phosphate aldolase ⟶ | + phosphate ⟶ + thymine | thymidine phosphorylase precursor ⟶ + | + phosphate ⟶ + | (none)
Related pathways
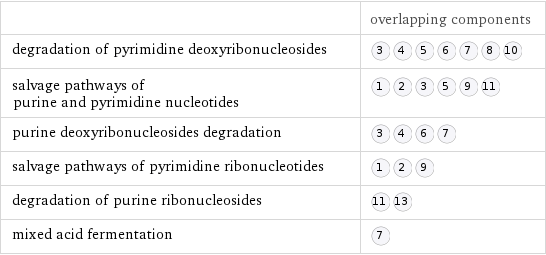
| overlapping components degradation of pyrimidine deoxyribonucleosides | salvage pathways of purine and pyrimidine nucleotides | purine deoxyribonucleosides degradation | salvage pathways of pyrimidine ribonucleotides | degradation of purine ribonucleosides | mixed acid fermentation |